Python#
In this little exercise, we will use Python in GRASS GIS to automatise the calculation of sun exposure for several times of the day. This was used within the HEAL project to model solar exposure of streets in Heidelberg so it could be integrated into the heat-avoiding routing in the HEAL app.
Start GRASS GIS and open your mapset from assignment 3.
Open the Simple Python editor.
Fig. 10 Open the Simple Python editor in GRASS GIS#
It contains some default Python code. Run the script by clicking the green arrow in the toolbar. What happens?
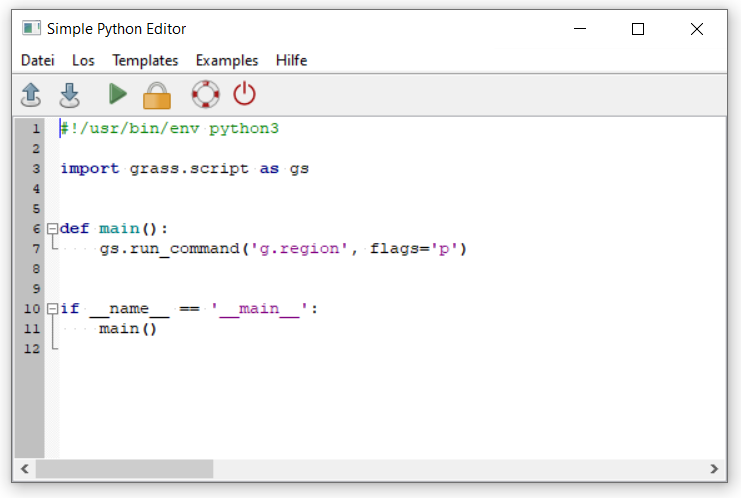
Fig. 11 Open the Simple Python editor in GRASS GIS#
Add a new line containing the
r.sunmaskcommand to calculate a shadow mask for 4 pm on June 21st.Warning
Make sure to add 4 whitespaces at the start of each line, so they align with the
gs.run_command('g.region', flags='p')command. This is important so that the code is executed correctly!#!/usr/bin/env python3 import grass.script as gs def main(): gs.run_command('g.region', flags='p') gs.run_command('r.sunmask', hour=h, ...) # Add all parameters here if __name__ == '__main__': main()
Use a for loop to calculate the shadow mask for 5 different times during the day.
By setting the parameter
hour=hin ther.sunmaskcommand, the sun mask will be calculated for different hours in each iteration of the loop.The name of the output file is defined by the parameter
output. This name needs to be adapted for each iteration of the loop. Setting theoutputparameter tof'sunmask_{h}'will do this for us. So do not change this.#!/usr/bin/env python3 import grass.script as gscript def main(): hours_of_day = [7, 10, 13, 16, 19] for h in hours_of_day: print(h) gscript.run_command('r.sunmask', output=f'sunmask_{h}', hour=h, ...) # add other parameters if __name__ == '__main__': main()
Download 1m digital surface model from https://opengeodata.lgl-bw.de/#/(sidenav:product/15)
Fig. 12 Open data for Baden Württemberg#
Import the digital surface model into GRASS GIS.
Run the script again with the new data.
How does the script look like that was used in the HEAL project?#
For the curious ones, here is the full script that was used to model solar exposures for all of Heidelberg within the HEAL project. In addition to the solar modelling itself, the script contains a lot of data handling as well as pre- and postprocessing of the data.
Note
If you want to understand what is happening here, take a programming course in the summer term such as the Python Introduction or the Geoscripting seminar.
#!/usr/bin/env python3
import rasterio as rio
from pathlib import Path
import subprocess
import glob
from datetime import timedelta
from dateutil import parser
import grass.script as gs
import logging
import numpy as np
logging.basicConfig(level=logging.INFO)
def find_neighbouring_tiles(tile_name: str):
""" Find the neighbouring tiles of a given tile.
:param tile_name: The name of the tile
"""
# Search for neighbouring tiles via ID
parts = tile_name.split('_')
parts[-1] = '*.tif'
id1 = int(parts[2])
id2 = int(parts[3])
ids1 = [id1 - 1, id1, id1 + 1]
ids2 = [id2 - 1, id2, id2 + 1]
file_names = []
for i in ids1:
for j in ids2:
parts[2] = str(i)
parts[3] = str(j)
file_names.append("_".join(parts))
return file_names
def main():
# Input: DOM Tile with ID
dsm_dir = Path('/Users/cludwig/Development/heal/data/amtliche_daten/DOM/')
dem_dir = Path('/Users/cludwig/Development/heal/data/amtliche_daten/DGM/')
output_dir = Path('/Users/cludwig/Development/heal/data/shadow_index/')
temp_dir = Path('/Users/cludwig/Development/heal/data/temp/')
dsm_files_txt = Path('/Users/cludwig/Development/heal/heal-shadow/grass/dsmfiles/dsm_files_sub2.txt')
doy = 240
# Read txt file without ending newline
with open(dsm_files_txt, 'r') as f:
dsm_files = f.read().splitlines()
for dsm_file in dsm_files:
logging.info(f'Processing {dsm_file}')
tile_name = Path(dsm_file).name
tile_stem = Path(dsm_file).stem
# Create virtual raster with neighbouring tiles
raster_files = []
for f in find_neighbouring_tiles(tile_name):
res = glob.glob(str(dsm_dir / f"**/{f}"), recursive=True)
if len(res) == 1:
raster_files.append(res[0])
elif len(res) == 0:
logging.warning(f"File {f} not found.")
vrt_file = temp_dir / tile_name.replace('.tif', '_neighbours.vrt')
cmd = ['gdalbuildvrt', str(vrt_file)] + raster_files
subprocess.call(cmd)
with rio.open(dsm_file) as src:
dsm = src.read(1)
dsm_meta = src.meta
xmin = dsm_meta['transform'][2]
xmax = dsm_meta['transform'][2] + dsm_meta['width'] * dsm_meta['transform'][0]
ymax = dsm_meta['transform'][5]
ymin = dsm_meta['transform'][5] + dsm_meta['width'] * dsm_meta['transform'][4]
xmin_buffered = xmin - 25
xmax_buffered = xmax + 25
ymin_buffered = ymin - 25
ymax_buffered = ymax + 25
dsm_file_buffered = temp_dir / tile_name.replace('.tif', '_buffered.tif')
subprocess.call(
['gdalwarp', '-overwrite', '-s_srs', 'EPSG:25832', '-t_srs', 'EPSG:25832', '-tr', str(dsm_meta['transform'][0]),
str(dsm_meta['transform'][4]), '-r', 'near', '-te', str(xmin_buffered), str(ymin_buffered), str(xmax_buffered), str(ymax_buffered), '-tap', '-te_srs',
'EPSG:25832', '-of', 'GTiff', vrt_file, dsm_file_buffered])
# Find DEM file
tilename_parts = tile_name.split('_')
tilename_parts[0] = 'dgm1'
tilename_parts[-1] = '*.xyz'
dem_tile_name = "_".join(tilename_parts)
dem_file = glob.glob(str(dem_dir / f"**/{dem_tile_name}"), recursive=True)
if len(dem_file) == 1:
logging.info(f'Reading {dem_file[0]}')
dem_file = dem_file[0]
elif len(dem_file) == 0:
logging.warning(f"File {dem_tile_name} not found.")
with rio.open(dem_file) as src:
dem = src.read(1)
dem_meta = src.meta
if dsm_meta['transform'] != dem_meta['transform']:
logging.info('transforming')
dem_file_aligned = temp_dir / Path(dem_file).name.replace('.xyz', '_aligned.tif')
xmin = dsm_meta['transform'][2]
xmax = dsm_meta['transform'][2] + dsm_meta['width'] * dsm_meta['transform'][0]
ymax = dsm_meta['transform'][5]
ymin = dsm_meta['transform'][5] + dsm_meta['width'] * dsm_meta['transform'][4]
subprocess.call(['gdalwarp', '-overwrite', '-s_srs', 'EPSG:25832', '-t_srs', 'EPSG:25832', '-tr', str(dsm_meta['transform'][0]), str(dsm_meta['transform'][4]), '-r', 'near', '-te', str(xmin), str(ymin), str(xmax), str(ymax), '-tap', '-te_srs', 'EPSG:25832', '-of', 'GTiff', dem_file, dem_file_aligned])
with rio.open(dem_file_aligned) as src:
dem = src.read(1)
dem_meta = src.meta
# Perform the shadow modelling using the buffered raster
year = 2024
hours = [10, 13, 16, 19]
date_str = f"{year}-01-01"
date_obj = parser.parse(date_str)
date_res = date_obj + timedelta(days=doy - 1)
minute = 0
timezone = 2
gs.run_command('r.import', input=str(dsm_file_buffered), output='dom', overwrite=True)
gs.run_command('g.region', flags='p')
gs.run_command('g.region', raster='dom')
gs.run_command('g.region', flags='p')
for hour in hours:
logging.info(f'Calculating shadows for {hour}:00...')
shadow_covered_file = output_dir / f'shadow&covered_{tile_stem}_{str(doy)}_{str(hour)}{minute:02d}.tif'
if shadow_covered_file.exists():
logging.info(f'File {shadow_covered_file} already exists. Skipping...')
continue
gs.run_command('r.sunmask', elevation='dom', output='shadow', year=year, month=date_res.month, day=date_res.day, hour=hour, minute=minute, timezone=timezone, overwrite=True)
outfile = temp_dir / f'shadow_{tile_stem}_{str(doy)}_{str(hour)}{minute:02d}_buffered.tif'
gs.run_command('r.out.gdal', input='shadow', output=str(outfile), format='GTiff', overwrite=True)
# Clip the virtual raster to the extent of the central tile
outfile_clipped = output_dir / outfile.name.replace('_buffered.tif', '.tif')
subprocess.call(
['gdalwarp', '-overwrite', '-s_srs', 'EPSG:25832', '-t_srs', 'EPSG:25832', '-tr', str(dsm_meta['transform'][0]),
str(dsm_meta['transform'][4]), '-r', 'near', '-te', str(xmin), str(ymin), str(xmax), str(ymax), '-tap', '-te_srs',
'EPSG:25832', '-of', 'GTiff', outfile, outfile_clipped])
# Fill nans in shadow mask
with rio.open(outfile_clipped) as src:
shadow = src.read(1)
shadow_meta = src.meta
shadow_filled = np.where(shadow == 255, 0, shadow)
with rio.open(outfile_clipped, 'w', **shadow_meta) as dst:
dst.write(shadow_filled, 1)
# Calculate difference between dom and dsm
height_diff = dsm - dem
covered_area = np.where(height_diff >= 2, 1, 0)
shadow_covered = np.where((shadow_filled == 1) | (covered_area == 1), 1, 0)
with rio.open(shadow_covered_file, 'w', **shadow_meta) as dst:
dst.write(shadow_covered, 1)
if __name__ == '__main__':
main()
